Disease Course Sequencing with the Event Based Model#
by Neil Oxtoby and Vikram Venkatraghavan
Disease Course Sequencing with Event-Based Modelling
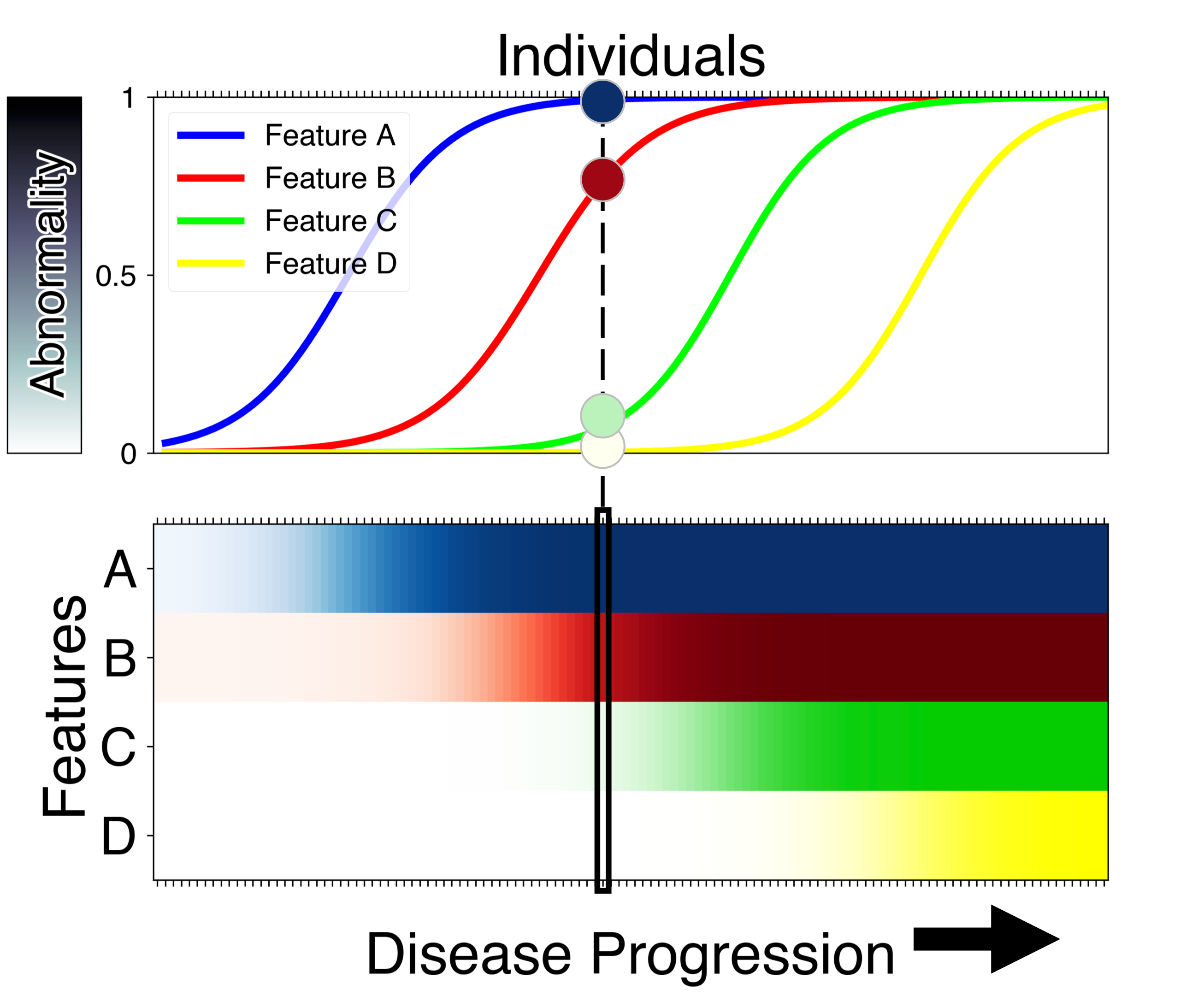
Event-Based Modelling (EBM) is a class of mathematical models, with associated Python softwares, that estimate a quantitative signature of disease progression (a Disease Course Sequence) using either cross-sectional or longitudinal medical data.
The softwares:
reconstruct the pathophysiological cascade (fine-grained temporal sequence of events) for a chronic, progressive disease
stages individuals along this fine-grained Disease Course Sequence, representing their cumulative abnormality along the group-average progression
does this all probabilistically and without predefined biomarker cutpoints
Software Python package Open source Tutorials
The software for classical EBM is distributed via the UCL POND group’s GitHub account, typically under the MIT license.
The software for the state-of-the-art Discriminative EBM as well as classical EBM (pyebm) is distributed via Vikram’s GitHub account, typically under the GNU General Public License v3.0.
Note: The classical EBM implementation in the two repositories differ in the type of mixture modelling (an essential step in EBMs) they support. The UCL POND group’s implementation supports KDE based mixture modelling as well as Gaussian mixture modelling (GMM), whereas pyebm only supports GMM.
The softwares should operate across operating systems, but specific requirements, e.g., python package versions, are detailed in each repository.
Usage#
The KDE EBM package includes user-friendly functions to perform key operations in the Disease Course Sequencing pipeline:
kde_ebm.mixture_model: fit_all_kde_models(...), fit_all_gmm_models(...)Converts multimodal biomarker data into event probabilities by fitting mixture models to patient/control data using Kernel Density Estimation or Gaussian mixture modelling.
kde_ebm.plottingPlotting tools for visualizing model outputs, e.g.,
mixture_model_grid(),mcmc_uncert_mat(),mcmc_trace(),stage_histogram()kde_ebm.mcmcTools for Markov Chain Monte Carlo fitting of the EBM, including bootstrap cross-validation.
The pyEBM toolbox can be used to fit a Discriminative EBM, or a classical EBM using Gaussian mixture modelling.
pyebm.debm.fit(...):Fits DEBM to multimodal biomarker data and inherently handles missing data. DEBM estimates average Disease Course Sequence for the entire cohort, any pre-defined subgroup of it, as well as for each subject in the cohort.
pyebm.ebm.fit(...):Fits EBM to multimodal biomarker data and inherently handles missing data. EBM estimates average Disease Course Sequence for the entire cohort.
Tutorial(s)#
We have developed an introductory tutorial to understand Disease Course Sequencing using Event-Based Modelling. In future, we will provide an example on real data from a publicly available dataset.
Tutorial 1: KDE EBM Hello World: example EBM on simulated data
This introduction to Event-Based Modelling is a walkthrough where you will fit an EBM using the KDE EBM software and simulated data.
Go to the tutorial 30 minutes cross-sectional data
Tutorial 2: pyEBM Hello World: example DEBM on simulated data
This introduction to Discriminative Event-Based Modelling is a walkthrough where you will fit a DEBM using the pyEBM software and simulated data.
Go to the tutorial 30 minutes crosssectional data
Wishlist.
Wishlist.
Installation#
KDE EBM installation is explained in the GitHub repository
pyEBM installation is via pip: pip install pyebm